The note is based on the contents of the introduction to SEM in R.
Table of contents
- 1. Basic concepts
- 2. Model without latent variables
- 3. Model with latent variables
- 4. Model evaluation: model fit index
1. Basic concepts
Structural equation modeling (SEM) is an umbrella term referring to a framework of linear models designed to explore relationships among both observable and unobservable variables.
1.1 Terms
- Observed vs. Latent:
- Observed variable: exists in the data (i.e. item/manifest variable)
- Latent variable: does not exist in the data (i.e. construct)
- Exogenous vs. Endogenous:
- Exogenous variable: not be explained by the model (like IV)
- Endogenous variable: be explained by the model (like DV)
- Indicator, factor, loading (terms in factor analysis):
- Indicator: an observed variable (i.e. item)
- Factor: an latent variable defined by indicators
- Loading: the path between an indicator and a factor
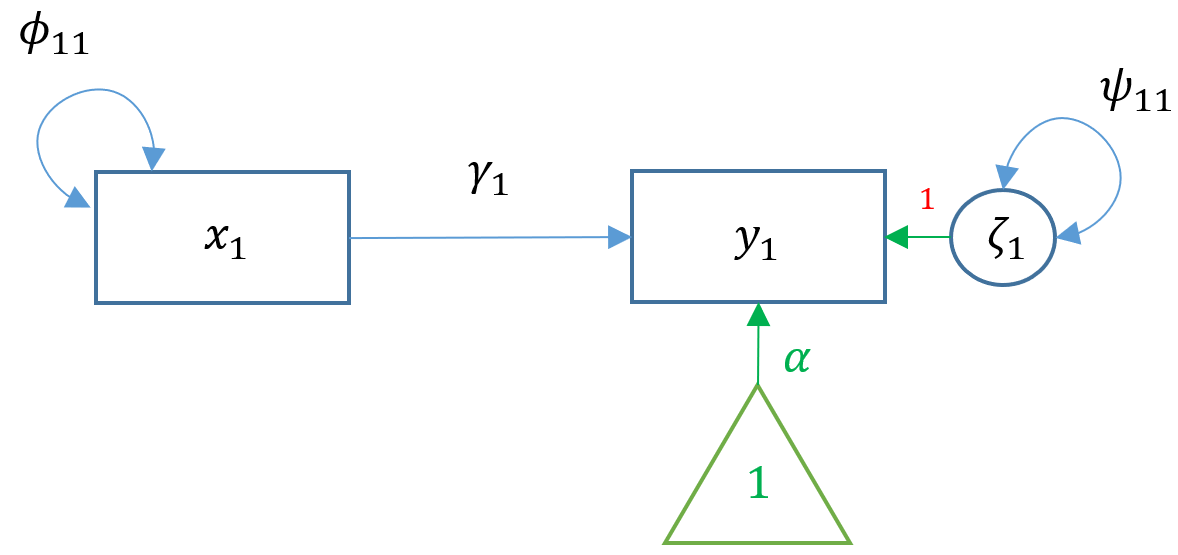
1.2 Path diagram

The following example is a structural model with two measurement models. Both exogenous variable (i.e. independent variable) and endogenous variable (i.e. dependent variable) are latent variables, which means that they are not derived directly from the data. Instead, they are constructed from some observable indicators respectively.

2. Model without latent variables
All exogenous and endogenous variables are observed.
2.1 Simple regression (1 ex & 1 en)
m1 = '
# regression
read ~ 1 + motiv
# variance (optional)
motiv ~~ motiv
'
m1 = sem(m1, data=dat)
print(summary(m1))Regressions:
Estimate Std.Err z-value P(>|z|)
read ~
motiv 0.530 0.038 13.975 0.000
Intercepts:
Estimate Std.Err z-value P(>|z|)
.read -0.000 0.379 -0.000 1.000
motiv 0.000 0.447 0.000 1.000
Variances:
Estimate Std.Err z-value P(>|z|)
motiv 99.800 6.312 15.811 0.000
.read 71.766 4.539 15.811 0.000- Intercepts:
.read: interceptmotiv: motiv’s mean
- Variances:
.read: residual variancemotiv: motiv’s variance
- Note:
lavaanuses MLE to do estimating;lmuses OLS instead.
2.2 Multiple regression (2+ ex & 1 en)

m2 = '
# regression
read ~ 1 + ppsych + motiv
# covariance
ppsych ~~ motiv
'
m2 = sem(m2, data=dat)
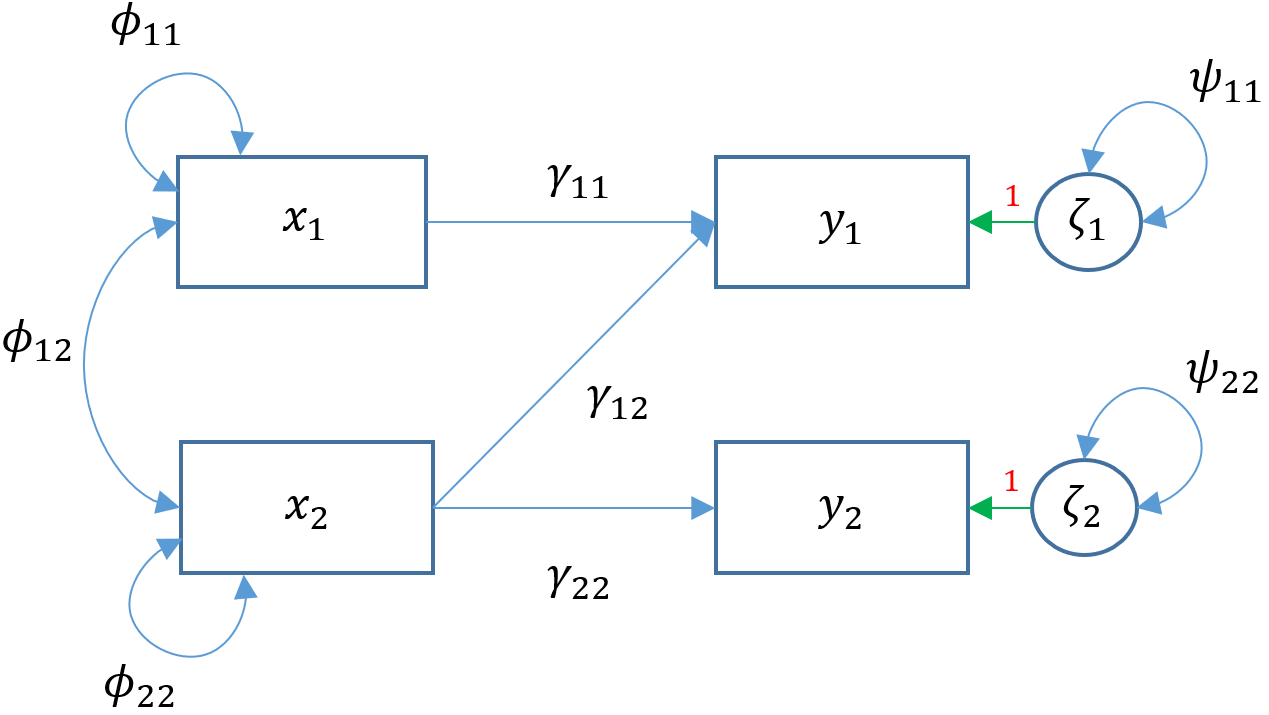
print(summary(m2))2.3 Multivariate regression (2+ ex & 2+ en)
Modeling multiple endogenous variables simultaneously:

m3a = '
# regressions
read ~ 1 + ppsych + motiv
arith ~ 1 + motiv
# covariance
# (lavaan estimates the covariance between endogenous variables by default)
ppsych ~~ motiv
'
m3a = sem(m3a, data=dat)
print(summary(m3a))Regressions:
Estimate Std.Err z-value P(>|z|)
read ~
ppsych -0.216 0.030 -7.289 0.000
motiv 0.476 0.037 12.918 0.000
arith ~
motiv 0.600 0.036 16.771 0.000
Covariances:
Estimate Std.Err z-value P(>|z|)
ppsych ~~
motiv -24.950 4.601 -5.423 0.000
.read ~~
.arith 39.179 3.373 11.615 0.000
Intercepts:
Estimate Std.Err z-value P(>|z|)
.read -0.000 0.361 -0.000 1.000
.arith -0.000 0.357 -0.000 1.000
ppsych -0.000 0.447 -0.000 1.000
motiv 0.000 0.447 0.000 1.000
Variances:
Estimate Std.Err z-value P(>|z|)
.read 65.032 4.113 15.811 0.000
.arith 63.872 4.040 15.811 0.000
ppsych 99.800 6.312 15.811 0.000
motiv 99.800 6.312 15.811 0.000Turn off the estimation of covariance between endogenous variables:
m3d = '
# regression
read ~ 1 + ppsych + motiv
arith ~ 1 + motiv
# covariance
ppsych ~~ motiv
read ~~ 0 * arith
'The above two models are not saturated models because the reports from lavaan show that there are remaining degrees of freedom (i.e. df ≠ 0): df(m3a)=1, df(m3d)=2. Add all possible parameters about endogenous variables to make the model saturated - df(m3e)=0:

m3e = '
# regression
read ~ 1 + ppsych + motiv
arith ~ 1 + ppsych + motiv
# covariance
ppsych ~~ motiv
'2.4 Path analysis (Multivariate reg & en ~ en)
Multivariate regression is a special case of path analysis, which allows an endogenous variable to be explained by other endogenous variables:

m4a = '
# regression
read ~ 1 + ppsych + motiv
arith ~ 1 + motiv + read
# covariance
ppsych ~~ motiv
'This model is not saturated with a df=1, which means you have left room to work with for adding parameters to make your model more complex. modindices function helps you decide add what:
print(modindices(m4a, sort=TRUE)) lhs op rhs mi epc sepc.lv sepc.all sepc.nox
20 arith ~ ppsych 4.847 0.068 0.068 0.068 0.068
17 arith ~~ ppsych 4.847 6.368 6.368 0.100 0.100
22 ppsych ~ arith 4.847 0.158 0.158 0.158 0.158
25 motiv ~ arith 4.847 0.632 0.632 0.632 0.632
14 read ~~ arith 4.847 16.034 16.034 0.314 0.314
19 read ~ arith 4.847 0.398 0.398 0.398 0.398
18 arith ~~ motiv 4.847 25.472 25.472 0.402 0.402mi is called 1-degree chi-square statistic, which means the model chi-square will drop how much after adding the term. However, be careful to change the model specification unless you have enough evidence from theories.
3. Model with latent variables
3.1 Measurement model (factor analysis)
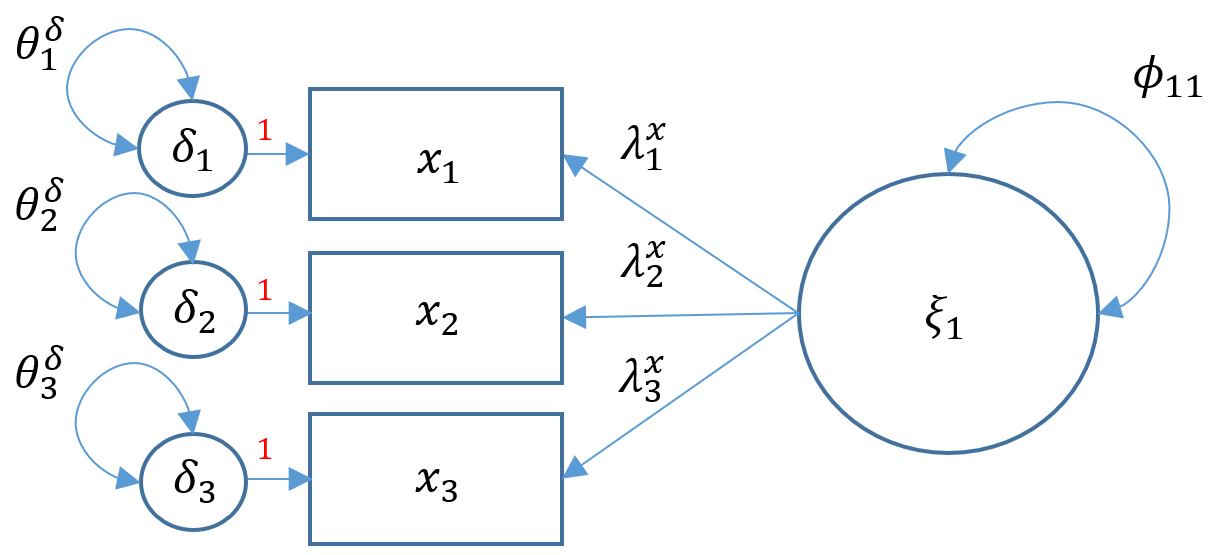
m5a = '
# indicator equation: factor =~ indicators
risk =~ verbal + ses + ppsych
# intercepts (by default no intercepts)
verbal ~ 1
ses ~ 1
ppsych ~ 1
'
m5a = sem(m5a, data=dat)
print(summary(m5a, standardized=TRUE) )lavaan 0.6.17 ended normally after 37 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 9
Number of observations 500
Model Test User Model:
Test statistic 0.000
Degrees of freedom 0
Parameter Estimates:
Standard errors Standard
Information Expected
Information saturated (h1) model Structured
Latent Variables:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
risk =~
verbal 1.000 6.166 0.617
ses 1.050 0.126 8.358 0.000 6.474 0.648
ppsych -1.050 0.126 -8.358 0.000 -6.474 -0.648
Intercepts:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.verbal 0.000 0.447 0.000 1.000 0.000 0.000
.ses -0.000 0.447 -0.000 1.000 -0.000 -0.000
.ppsych -0.000 0.447 -0.000 1.000 -0.000 -0.000
Variances:
Estimate Std.Err z-value P(>|z|) Std.lv Std.all
.verbal 61.781 5.810 10.634 0.000 61.781 0.619
.ses 57.884 5.989 9.664 0.000 57.884 0.580
.ppsych 57.884 5.989 9.664 0.000 57.884 0.580
risk 38.019 6.562 5.794 0.000 1.000 1.000- In order to identify a factor model with three or more items:
- marker method fixes the first loading of each factor to 1.
- variance standardization method fixes the variance of each factor to 1 but freely estimates all loadings.
- By default, lavaan reports the results of the marker method (1.000, 1.050, -1.050); by requesting the standardized version, lavaan reports the results of the variance standardization method in
Std.allcolumn (0.617, 0.648, -0.648).
3.2 Structural Model
Two steps:
- First, construct measurement models.
- Second, specify how the latent variables relate to each other.
This is a structural model with two endogenous variables (i.e. a mediation analysis model):

m6b = '
# measurement models
adjust =~ motiv + harm + stabi
risk =~ verbal + ses + ppsych
achieve =~ read + arith + spell
# regression
adjust ~ risk
achieve ~ adjust + risk
'
m6b = sem(m6b, data=dat)
print(summary(m6b, standardized=TRUE, fit.measures=TRUE))4. Model evaluation: model fit index
In regression analysis, there are four common criteria for evaluating a model: 1) adjusted R2; 2) MSE(mean square error); 3) RSE(residual standard error); and 4) information criterion(AIC & BIC). In SEM, there are also four commonly used indexes for model evaluation. Here are some important points to remember:
- The model fit indexes only have true meanings when the model’s degree of freedom is greater than 0 (i.e. not a saturated model).
- The model fit indexes are test statistics with a null hypothesis stating, the model fits the data well. Specifically, this null hypothesis implies that the model-implied covariance matrix is identical to the population covariance matrix .
- Hence, rejecting the null hypothesis means that our model does not fit the data well.
Request lavaan for additional fit statistics:
summary(model, fit.measures=TRUE)4.1 Model chi-square
- The chi-square value increases when there is a larger difference between the current model-implied covariance matrix and the sample covariance matrix - more probable to reject H0.
- When Model Test User Model: P-value (Chi-square) < 0.05, reject H0.
- 👎 Drawback: the chi-square is extremely sensitive under large samples. You may always get a significant p-value when sample size is greater than 400. In this case, you should refer to other indexes.
4.2 CFI (Comparative Fit Index)
It is a relative fit index - comparing your model with the saturated model based on a baseline model (just like the test of goodness of fit in logistic regression - see more details):
“An relative fit index assesses the ratio of the deviation of the user model from the worst fitting model (a.k.a. the baseline model) against the deviation of the saturated model from the baseline model.”
- The closer the CFI is to 1, the better the fit of the model.
- Empirically, CFI greater than 0.90 (or 0.95) indicates a good fit.
4.3 TLI (Tucker Lewis Index)
It is also a relative fit index:
- The closer the TLI is to 1, the better the fit of the model.
- Empirically, TLI greater than 0.90 indicates a good fit.
- TLI and CFI are highly correlated so only one of the two should be reported.
4.4 RMSEA
It is not a relative fit index. Instead, it is a absolute index which does not involve comparisons. Some rules of thumb:
- → good fit
- but → reasonbale fit
- → poor fit